GFDL Runtime Environment: The FMS Runtime Environment (FRE)
From the perspective of model execution and post- processing, run-time environments have emerged to assist users in configuring, compiling, and running models, as well as post-processing and archiving output data files. They are environments intended to reduce the time to get the model up and running and at the same time reduce errors caused by manual manipulation of configuration files. GFDL was an early adopter of such environments and the management of climate model runs is done using FMS Runtime Environment (FRE), a framework that encapsulates complete model configurations in a single file, from submission of model runs, archiving model outputs, to post-processing the output data and creating automated analysis figures. FRE has been managing the bulk of GFDL’s climate runs since 2004. The FMS experiments are described in a comprehensive XML configuration file–from source code checkout to post-processing of output data. This XML file is processed by the runtime environment which generates the appropriate scripts for executing the model. The environment also generates scripts, and their dependencies, that are automatically scheduled for execution by the batch system.
One goal of FRE is to achieve reproducibility of a model run, a central element of the scientific method. The aim of this research is to enable a scientific query to be formulated, the answer to which points either to a model output dataset, or a model itself, that can be run to provide the result, perhaps even as a web service.
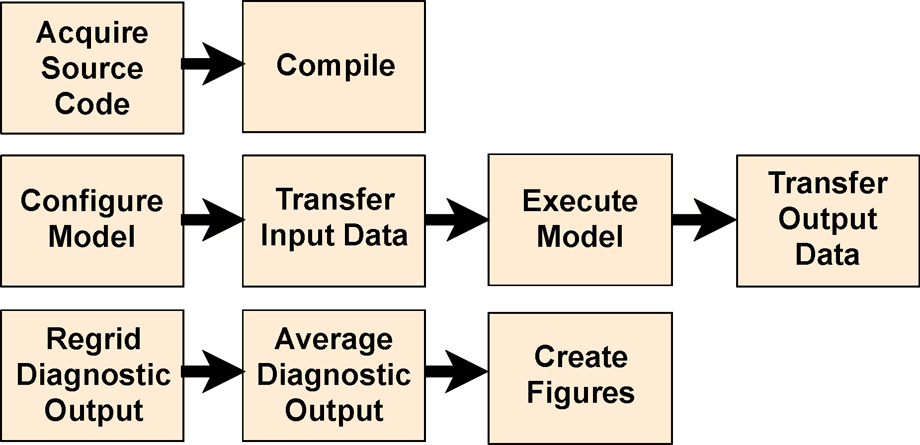
FRE is a set of workflow management tools that read a model configuration file and take various actions.


